CRISPRSuite
CRISPRSuite programs and tools currently consist of CRISPRTarget, CRISPRDetect, tracrRNAFinder and CRISPRBank. An overview is shown in Figure 1.
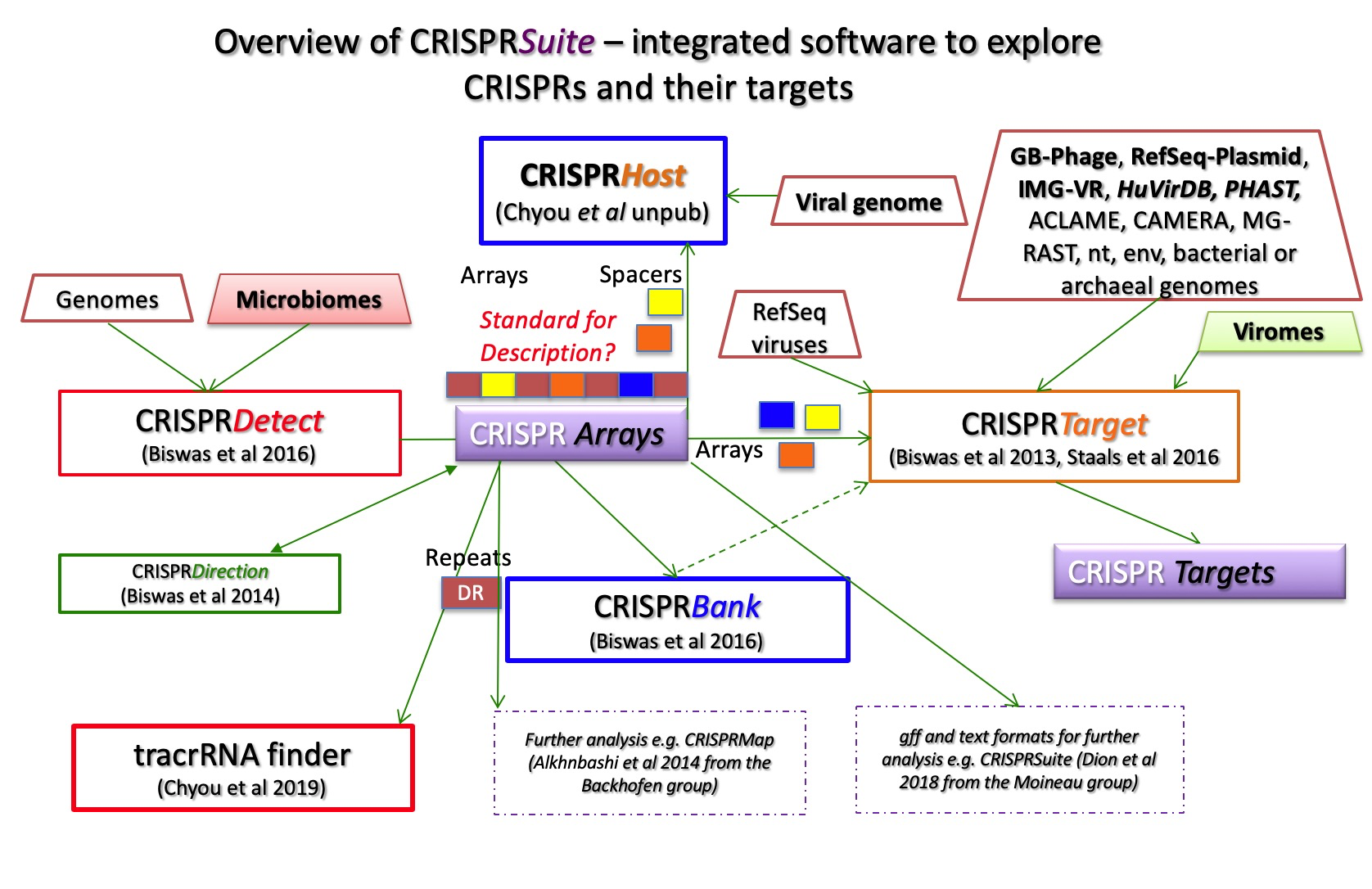
Common CRISPRSuite tasks
1. I want to know what the spacers in my arrays target
Use CRISPRTarget
(CRISPRTarget is the only tool for this purpose. Alternative generic approaches would be to try blastn, or water)
2. I have a genome or draft genome and want to find CRISPR noncoding RNAs
Use CRISPRDetect 2.4 (or download )
(Our recent publication outlines the advantages of CRISPRDetect, alternative approaches would be to use CRISPRFinder (2007), CRT (2007), Minced (2014), CRISPRCasFinder (2018), or CRISPI or CRISPRDisco.
3. I want to know if any other species have this repeat?
Use CRISPRBank or CRISPRDetect then CRISPRBank (RefSeq95 7/2019)
(Alternative: CRISPRCasdb)
4. I have predicted an array with CRISPRDetect or another tool and want to correct the direction/strand.
Use CRISPRDirection (for download) or repeat the analysis with CRISPRDetect which includes CRISPRDirection 2.0
5. I want to find tracrRNAs, use tracrRNA finder
Tools which provide functionality that CRISPRSuite currently does not have:
CRISPRMap- Given a repeat sequence classify it.
(please suggest additions to this list)
A community editable list of Bioinformatics tools for CRISPR Biology:
Creation 3/8/2016
Last update: 8/11/2020
By Chris Brown